Systems Biology||Synthetic Biology||Molecular Biophysics
Systems Biology and the Structure, Dynamics and Function of Global Gene Regulatory Networks
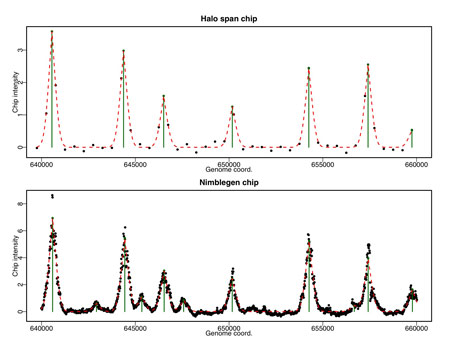
One of the major goals of Systems Biology is to decipher the structures and understand the dynamics of cellular networks in a way that allows us to predicatively rewire these cellular circuits for our own purpose. This ability will facilitate the rewiring of disease-perturbed networks back to health and the engineering of novel cellular factories. We are interested in utilizing the tools and frameworks of Systems Biology to learn more about how structure and dynamics in gene regulatory networks, those networks composed of transcription factors and cis-regulatory elements, lead to the emergence of complex phenotypes. We aim to take advantage of the growing number of fully sequenced microorganisms, their experimental tractability, high-throughput analytical technologies and computational modeling approaches to decipher structures of gene regulatory networks from across the tree-of-life. Understanding how Nature employs a diverse set of structures to satisfy its regulatory goals will greatly impact our ability to engineer these networks. We leverage numerous high-throughput technologies available to us in the UC Davis Genome Center, which include next-generation Solexa sequencing, microarray scanners, proteomics, metabolomics and bioinformatics. For more information on the Genome Center please visit: (http://genomics.ucdavis.edu/).
Engineering biological systems
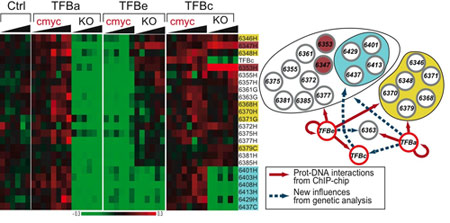
Physical associations within gene regulatory networks are principally composed of two types of interactions – protein-protein and protein-DNA. Small changes in the biophysical properties governing these interactions can have profound influence on the behavior or the network in question. We are interested in understanding the biophysical and biochemical principles that govern how these important associations contribute to network properties, how Nature leans on these evolutionary levers to build novel networks and how we can utilize this knowledge to engineer custom circuits that have application across biology. We are currently collaborating with Dr. Ling Yuan at the University of Kentucky, Lexington to engineer novel general transcription factors to help us better understand the structural and biophysical determinants of their association to DNA and other transcription factors.
Microbial Phenotypes
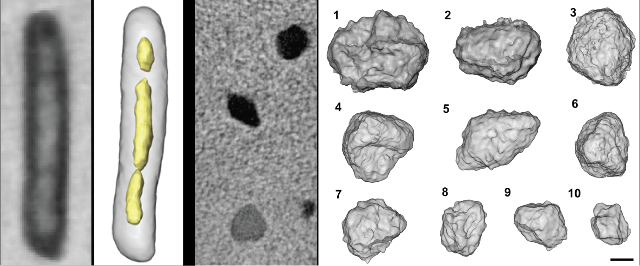
One goal of systems biology is to build models of cellular regulatory and physiological networks that can accurately predict cellular phenotypes. From the standpoint of genomic complexity, bacterial and archaeal systems are ideally suited for this endeavor. However, due to their physical dimensions, it is difficult to get numerous accurate quantitative phenotypes for microbial cells. Two types of phenotypes are typically measured for unicellular organisms (a) growth (b) morphology. Growth is relatively trivial to measure, easy to quantify and may provide a useful window into cellular metabolism. Morphology, however, is more difficult to routinely quantify due to the relatively small size of most bacteria and archaea. We are interested in using X-ray tomography, a novel imaging technique pioneered by our collaborators at the Lawrence Berkeley National Laboratory (http://ncxt.lbl.gov/) that provides resolution between that of light microscopy and electron microscopy, to quantify various aspects of cellular morphology in microorganisms. This technology provides distinct advantages over light microscopy (improved resolution) and electron microscopy (high-throughput and hydrated samples) that should make its continued development highly valued for numerous systems biology initiatives.