Systems Biology||Synthetic Biology||Molecular Biophysics
DNA sequencing technologies are providing incredible opportunities for discovery and understanding in the biological sciences. This is particularly true in the microbial sciences, where new tools now offer views into the genomes of many model and non-model microorganisms and into the metagenomic composition and dynamics of microbial communities. However, studies interpreting the functional expression of these genomes (what the genomes do and how they do it) are lagging far behind. We are interested developing experimental and computational approaches to bridge this knowledge gap.
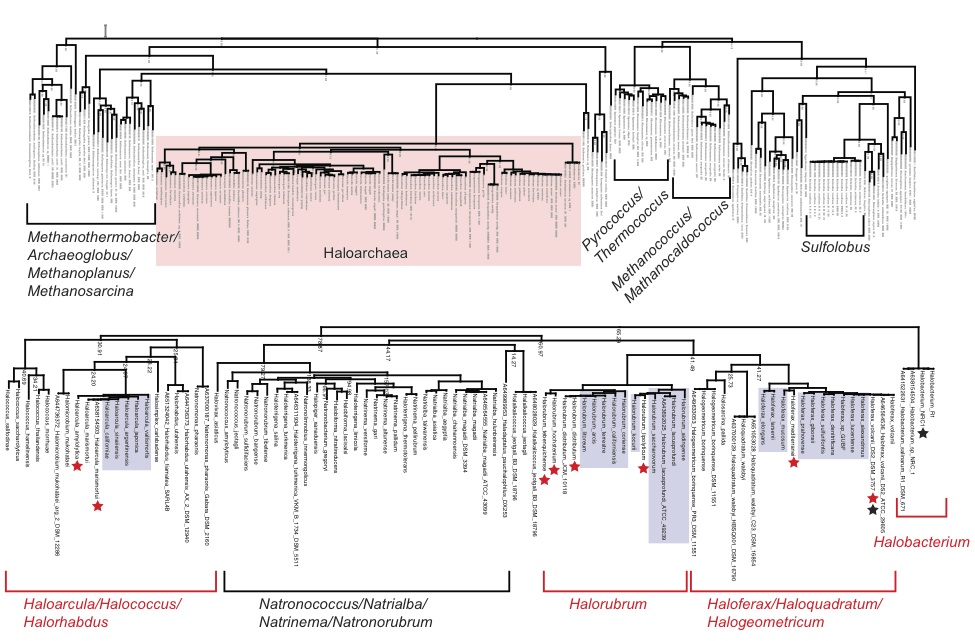
Sequencing Haloarchaeal Genomes
Collaboration with Profs. Jonathan Eisen (UCD) and Prof. Aaron Darling
Project Supported by NSF (EF-094953)
In 1977, Carl Woese catalyzed a revolution in our understanding of the living world by presenting evidence that there are three main branches on the tree of life and that one of these branches, the archaea, had been previously unrecognized. Despite the general acceptance of the existence of the archaea as a separate branch in the tree of life, they are still relatively poorly understood compared to bacteria and eukaryotes. This is not to say there have not been significant advances in our understanding of archaea, but rather that the work pales somewhat in comparison to studies of model bacteria such as Escherichia coli and Bacillus subtilis and many others, and model eukaryotes ranging from ciliates (e.g., Tetrahymena thermophila), to plants (e.g., Arabidopsis thaliana), to animals (e.g., Drosophila melanogaster, Caenorhabditis elegans, humans, mouse, Xenopus, etc). This is unfortunate since archaea represent one of the main lines of descent of all living organisms (and thus have a lot to tell us both about the generalities of biological processes as well as what novel processes might have originated in their history).
There is however one group of archaea that has enormous potential to serve as a model group for both biological and environmental studies. The group we refer is the family Halobacteriacea in the Phylum Euryarchaeota. In this project we aim to catalyze the further development of haloarchaea into model organisms through a high resolution genomic sequencing project. We have deposited genome sequences for >60 haloarchaeal isolates in NCBI and are busily analyzing the data therein.
Comparative Haloarchaeal Metabolomics
Collaboration with Prof. Oliver Fiehn (UCD) and Dr. Tobias Kind
For microbial species, the metabolome may be one of the ultimate and functionally relevant expressions of the genetic information encoded within their genomes. Indeed, understanding the complex relationship between the genome, the metabolic potential it encodes, and how it is regulated remains one of the exciting and mysterious challenges of modern biology. Moreover, genomes of both cultivated and uncultivated species likely represent a gold-mine of novel biochemistry and of small molecules that may be of pharmacological and/or biotechnological import. The archaea represent a class of organisms that epitomize the growing disconnect between our ability to sequence genomes and our knowledge of what those genomes encode. It is becoming “easy” to sequence these genomes but we typically know the function of less than half of the encoded genes (many of which can be reasonably assumed to have metabolic activity). As a consequence, we are collaborating with Drs. Oliver Fiehn and Tobias Kind to study the archaeal metabolome using liquid chromatography coupled to tandem mass spectrometery (LC-MS/MS), focusing initially on comparative analyses of haloarchaeal species.