| Structure, Dynamics and Function of Gene Regulatory Networks |
| One of the major goals of Systems Biology is to decipher the structures and understand the dynamics of cellular networks in a way that allows us to predicatively rewire these cellular circuits for our own purpose. This ability will facilitate the rewiring of disease-perturbed networks back to health and the engineering of novel cellular factories. We are interested in utilizing the tools and frameworks of Systems Biology to learn more about how structure and dynamics in gene regulatory networks, those networks composed of transcription factors and cis-regulatory elements, lead to the emergence of complex phenotypes. We aim to take advantage of the growing number of fully sequenced microorganisms, their experimental tractability, high-throughput analytical technologies and computational modeling approaches to decipher structures of gene regulatory networks from across the tree-of-life. Understanding how Nature employs a diverse set of structures to satisfy its regulatory goals will greatly impact our ability to engineer these networks. We leverage numerous high-throughput technologies available to us in the UC Davis Genome Center, which include next-generation Illumina sequencing, microarray scanners, proteomics, metabolomics and bioinformatics. For more information on the Genome Center please visit: (http://genomics.ucdavis.edu/). |
| Development of Archaeal ChIPSeq |
| Chromatin immunoprecipitation followed by next generation sequencing (ChIPSeq) is one of the most powerful technologies for investigating transcription factor-promoter associations in vivo. We have developed a robust experimental and informatic pipeline for conducting ChIPSeq experiments in the archaea. Our strategy utilizes chromosomal epitope tagging to investigate the dynamics of transcription factor activity. |
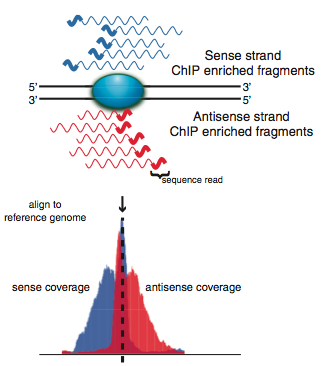 |
| Dynamics of Transcription Factor Binding |
| Gene regulatory networks are dynamic. Understanding their function requires that we understand the “rewiring” that occurs in GRNs in response to stimuli. We are currently applying ChIPSeq, quantitative proteomic, and transcriptional profiling approaches to study the dynamics of the Halobacterium salinarum GRN in response to heat shock. |
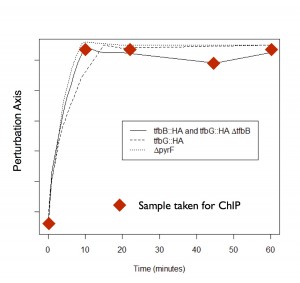 |
| Molecular Determinants of Paralogous Transcription Factor Cross-Talk |
| Gene duplication is one critical mechanism for the evolution of GRNs. This mechanism has important functional consequences that become readily evident using ChIPSeq – particularly the imperfect (overlapping) partitioning of promoter space by paralogous transcription factors. Through experimental and computational approaches, we are currently exploring some molecular-level mechanistic hypotheses regarding the activity of paralogous factors. |
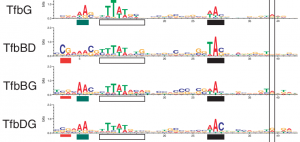 |



